XG (11-5): Reference for different inhibition mechanism from Prof. Segel's book "Enzyme kinetics-behavior and analysisi of repid equilibrium and steady-state enzyme sysems"
Enzyme kinetic_Ch 3_Segel IH.pdfInhibition mechanism for known sirtuin inhibitors.
PL (10/25): Attached is part of the information collected. More information and proposed discovery based on these mechanism will be updated soon.
Inhibition mechanism of known sirtuin inhibitors_part 1.docx
PL (10/28): An updated survey of sirtuins inhibition mechanism is attached. Knowing or assuming inhibition mechanism, especially those unusual modes, can help us find new class of inhibitors. RC (11-1):
Inhibition mechanism of known sirtuin inhibitors.docx
RC (11-1): Regarding your observation " In the crystal structures of SIRT3/peptide/piceatannol (4HD8) and SIRT5/peptide/resveratrol (4HDA), these molecules are found to cover the pepide binding pocket and may act by affecting the details of peptide binding. The ligand binding outside the regular binding pockets are certainly something not explored. [11]", please see my comments on the Sirtuin project 1 page which identifies some issues to look into, vis-a-vis our related interest in base exchange activators. I had already read this paper several months ago.
Proposed research on Sirtuin Project 10-21-2013
PL (10/21): In the attached Word document, I provided the workflow for proposed research. Proposed research_PL_XG.docx
Uciechowska, U. et al. Thiobarbiturates as sirtuin inhibitors: virtual screening, free-energy calculations, and biological testing. ChemMedChem 3, 1965–76 (2008)
and
Uciechowska, U. et al. Binding free energy calculations and biological testing of novel thiobarbiturates as inhibitors of the human NAD+ dependent histone deacetylase Sirt2. Med. Chem. Commun., 3, 167-173 (2012)
show examples of using MD and MD-based MM-GBSA and LIE calculations in the inhibitor discovery for SIRT2.
RC: Thanks for the overview. Following are my preliminary comments:
Project 1): -- a) Incorporate time required for experiments - needed for LIA. Let's get more specific about the timeline and when experiment and computation may be carried out in parallel. (If we need to scale up the experimental effort down the line, CJ could possibly help out with experiments on some molecules. But I don't believe that's necessary at this time.)
XG (10-22):
- We will start with the three molecules (ChemBridge 5281077, 4102009, 9147724), which are currently available in the lab. For LIE, IC50 value needs to be obtained from experiment. Specifically, the work needed for every compound: 1 week for study of inhibitor physiochemical properties. Depends on the solubility, another week for IC50 measurement (try 8-12 different inhibitor concentrations). Notice that, for IC 50 measurement, three molecules can be done together. Therefore, it will take 3weeks for lab to provide solid data.
- Ping pointed out since the bigger sampling and wide range of IC50 are important for LIE, we would expect to test more compounds which IC50 fall into nM range. Currently we have 5 compounds which IC50 in mM range, 4 compounds in uM range, 3 unknown. Early this year, GSK reported (Disch, JS et. al Journal of Medicinal Chemistry (2013) 56: 3666-3679.) that certain group of small molecules can have IC50s (SIRT3) 7-33 nM. We can use those compounds as seed to search some new molecules for lab to test.
- Recall the method Eric used to generate 10 molecules (I ordered 3 of them).
(2) Then used GlideXP docking to reduce the number to 57 (1st pass) to 25 (2nd pass). ChemBridge 5281077, 4102009, 9147724 are chosen from them.
- Ping will decide the methodology to provide new candidates. We may also need to decide how many more will be needed.
b) Will we try to use LIA for a predictive model without MD simulations as well?
PL: We will try LIA without MD, because it is not as time consuming.
c) What is the approximate time required for each MD simulation? This will affect the computation-experiment discovery workflow.
PL: Benchmark is needed. Depend on the computer system and model system, for current 16-cpu node, it may take a couple of days to get 1 ns of MD simulation alone, not including the time for parameterization and preparation. For a low budget GPU workstation shown here, http://ambermd.org/gpus/recommended_hardware.htm, we can achieve 1-2 ns of MD in a day depending on the setting and simulation code used.
Project 2):
a) Should be subdivided by inhibition mechanism; how many subprojects? This is one point I mentioned at the group meeting.
PL: The inhibitor for base exchange is the easiest, and for others, more time is needed for literature survey and planning.
b) Will each be treated as congeneric series? Will there be an attempt to develop a predictive model for each congeneric series based e.g. on LIA? Or do we feel that congeneric series assumptions will not hold for these mechanism-based models?
PL: The validity and applicability of LIA need to be tested first.
c) Regarding the proposed MD simulations: if these are not used for the generation of a fast predictive model, one has to consider whether it is more appropriate to do them at the lead discovery or lead optimization stage. Do you consider this step to be part of lead discovery? That appears to be the case given that there are no experiments in the workflow prior to MD. There are obvious advantages to MD for lead optimization (see below) whereas for lead discovery when the molecules can all be purchased without synthesis, one needs to show that computation will be faster than experiment.
PL: There are many tests need to done, for MD as well. One method/protocol may not be enough to generate the new leads we are looking for, and we may need to broaden our search to consider other possibilities. And we also want to confirm that the method/protocol doesn't produce too much false positives. Right now, many assumptions are all up in the air. My strategy is to begin with the easiest/simplest approach, and experimental test can be use to validate the approach, and computational approaches can be used to cross-examine each other. For example, from the docking tests I run so far, it appears more conformation sampling is needed for receptor to take into account of the flexibility and ability to accommodate certain ligands. However, do we need to generate a larger pool of receptor conformations, do we need to refine (MD, annealing,minimization, etc) the structural obtained? Or if we should settle for the best scores to avoid the computational cost? I recently run a analysis on apo-enzyme using elastic network model (ENM), and it shows that the two domains can undergo slow breathing motions to open up NAD+ and acetylated peptide binding pockets to accommodate larger ligands. Although multiple crystallographic structures have shown such changes, but enable such dynamics motion may allow us to get more potential hits.
RC: As part of this project, it would be nice to mention all the different modes of inhibition that we have identified (since it appears no one has really discussed these different modes in one paper), prior to focusing on one particular class of inhibitor.
RC: To what extent will you need to use additional compute resources for virtual screening and MD? Would you like to use our Beowulf cluster for this? Will you need to reconfigure/install new software on the cluster as a sysadmin to do so and if so, when would you do this?
PL: My plan is to first use the available resource for smaller tests. If there is such need for more computing resources, we can discuss what is the best option. I have already install all the software needed for MD simulation under my own directory, including NAMD for MD, AmberTools for preparation and analysis, Firefly for QM calculations, I also install openmpi in case MPI is used for certain programs. However, I don't see much gains using MPI cross nodes. These nodes are not configured equally. More storage/data server may be needed if we do decide to run MD.
RC: Regarding the workflow for lead discovery and optimization, I have had detailed discussions on how this is done in leading academic and industrial drug discovery groups. FEP is most commonly used in the lead optimization phase. Typically, they "grow" drug structures within the active site starting from a hit identified in the lead discovery phase. "Mutations" are made on the lead structure, based on common functional group substitutions and inspection of possible relevant interactions. As part of the lead optimization procedure, after several iterations, the compound is synthesized (we may not be able to do this now due to need for synthetic chemistry) and the crystal structure solved with collaborators (often a CRO). They claim the latter is essential to be able to validate the computational optimization approach. I am assuming that the above project outlines do not consider lead optimization, and our immediate next papers will focus on lead discovery protocols suitable for a high quality journal. Two groups I have spoken to at length are the Jorgensen group and the DE Shaw / Schrodinger / Nimbus group. I have one colleague at DE Shaw who wrote much of Desmond, which is used in the FEP calculations. I have two colleagues at Schrodinger/Nimbus who have been heavily involved with using Desmond in FEP calculations for lead optimization. They have recently sold a lead to the pharma industry. Jorgensen uses MC for FEP sampling, whereas Schrodinger/Nimbus uses MD with Desmond, and claims the latter is much faster. I have asked these colleagues if they would be willing to speak with the PMC-AT group regarding their approaches, and they may be available to do so. It could be helpful for us to determine whether we would do such lead optimization work ourselves possibly using their tools or through a collaboration with them. PL, please let me know if/when such discussion would be useful to us. Perhaps we should first identify good leads. XG, please let me know what you have found about crystallography via CROs. I also have a colleague who is in the management of a CRO (or an analogous academic organization called ARO - apparently there is one at Duke) that handles crystallography. I will provide you with her email address. If/when we have specific questions, you can compose an email to this colleague with cc to me to learn more. Let me know.
PL: FEP was a preferred choice of method in the past, but it usually requires more simulations (depends on number of windows) than MM-PB(GB)SA with MD (single or up to three simulations). I will certainly be interested to discuss with colleagues working in this field, to know better about their methods of choice and their strategy for drug discovery.
RC: Note that I am distinguishing between lead discovery and optimization. We need to characterize the objective of our next papers as one or the other - I think we are proposing lead discovery as the primary focus. One reason I mention this is that lead optimization is the subject of intense academic and industrial efforts as mentioned above and our efforts would be judged in the context of these state-of-the-art methodologies.
XG(10-22): A quotation was submitted to Genscript for the cost of "CrystalPro Structural Biology Service". The cocrystal structure will include HSIRT3/Carba-NAD+/peptide/AC93253. The cost will cover the x-ray data and structural file. Wait for response.
The current questions for CRO are time frame and cost. It will be nice to have other resource to request a quote for price comparison.
RC: Is the service you mentioned above from a CRO?
XG: GenScript is a biotech company which provide differet kinds of product (protein purification kit, PCR cling kit...) and services (like customized peptide snythesis, Animal model, even lead optimization services).
RC: Did they provide any estimate of when they will get back to us?
XG: Normally 2-3 business days.
XG(10-25): Quote from GenScript:
Phase 1. Protein Expression
Building expression construct, small scale expression and optimization, then perform 1L pilot expression and one step purification - $1,800
5L large scale expression and 3 steps purification - $2,700
Estimated total price - $4,500; Turnaround time 8 weeks
*The goal is to obtain 3-10 mg of protein at >90% purity for crystallography. Depending on the yield, larger scale might be needed to obtain sufficient proteins. And tag removal step will also be charged separately.
Phase 2. Protein crystallization
Crystallization Screening and optimization of conditions
Estimated price - $5,000; Turnaround time 1-4 weeks
Phase 3. Structure determination
Collection of high-resolution X-ray data using synchrotron source, and structure determination with crystallography software
Estimated price - $5,500; Turnaround time 4-7 weeks
Prepared by Kevin Shi, Ph.D.
Contact info:
Inside Sales Manager
GenScript USA Inc.
860 Centennial Ave. Piscataway, NJ 08854, USA
RC (11-4): Is this for just one structure? If we want to solve a number of complexes of the same protein with different drugs, would this be charged separately under Phase 2? For example, we could request crystallization of published complex along with known crystallization conditions, along with a new complex from our screening, at the same time.
How does the protein obtained from phase 1 differ from what we use now - in quantity, purity or both?
Please consider what published complex we should choose, and as we screen new molecules, think about which new complex would be best to show crystallographic data on, as part of the next paper.
XG (11-4): Yes. This is for one cocrystal structure (Enzyme + NAD+ +Substrate + small compound. With different drugs, this will be charged separately from Phase 2 and after. Both crystallography and kinetic study require high purity of enzyme. The crystallography asks for high amount of protein to be concentrated enough to crystallize.
RC: Is this the only CRO we have been able to solicit?
XG: Yes.
RC: Ok, so you have not been able to find a list of CROs with contact info.
RC: I can forward background info on the requested crystallography to the CRO contact I have if it is sufficiently detailed and in a proper format.
XG: We need to decide which cocrystal structure we are interested in. We also need to know what kind of service they can provide, such as do they synthesize cocrystal?do we need to provide protein? If so, the amount, in liquid phase or need to be lyophilized? ...
RC: Ok please compose an email with the background info and questions and provide to me when ready. You should also think about where in the workflow this would be used.
XG(10-25): A general quote request is drafted. I also prepared a template for quote. What we are interested currently, are the cost and turnaround time for those breakdown services (protein expression and purification/ crystallization / structure determination). The first round email will provide us a rough idea about the CRO without getting into details of our projects. More specific questions can be asked after the general quote is acceptted. Email for CRO outsource service.docxQuote template for general info for outsource crystallography services.docx
RC: We should aim to choose a project for the next paper, along with a detailed schedule of tasks for such paper, within the next week. We should also have an idea of the type of journal we will submit to (e.g. med chem vs biochem/comp bio journals). I have a slight preference for project 1 to start. Can you provide a publication that is closest in layout to the paper we might consider writing?
PL: We can start with both project 1 and part of project 2 (the inhibitor discovery toward base exchange reaction), because the extra information we obtain from project 2 will help us understand better the success and failure of the method/protocol we use in project 1.
For project 1, we can set our target on PLoS Computational Biology, similar to the one published by Matthew P. Jacobson.
Dolghih, E., Bryant, C., Renslo, A. R. & Jacobson, M. P. Predicting binding to p-glycoprotein by flexible receptor docking. PLoS Comput. Biol. 7, e1002083 (2011)
Or ACS Med. Chem. Lett and ChemMedChem, similar to the following articles,
Kohlmann, A., Zhu, X. & Dalgarno, D. Application of MM-GB/SA and WaterMap to SRC kinase inhibitor potency prediction. ACS Med. Chem. Lett. 3, 94–99 (2012)
Uciechowska, U. et al. Thiobarbiturates as sirtuin inhibitors: virtual screening, free-energy calculations, and biological testing. ChemMedChem 3, 1965–76 (2008).
The later two is probably better suited for project 2.
RC: Ok please provide a detailed schedule for project 1 per my questions above with attention to the interplay between experiment and computation and when these will be done in series/parallel, what info will be passed between experiment and computation. After this week we can start on this task list. (Then we can do the same for project 2, including the breakdown by inhibitor mode described above.)
Please post the papers here or on dropbox (not just citations); please ask XG for info on dropbox folder if needed.
RC: Please also indicate the novelty with respect to existing literature of each of the proposed project strategies. We have discussed several of these but they should be summarized here for convenience. In particular, the novelty of the activator design project is obvious - and this is one reason this is an attractive project. The drug industry has not established systematic methods for activator design. Lead optimization is fundamentally different than for inhibitors - it does not involve optimization of binding affinity, but rather optimization of catalytic turnover based on a kinetic model. There is a good chance that, if successful, future work on this topic could eventually be published in high impact journals like PNAS. (An important experimental part of this project, as XG mentioned, is to determine through study of base exchange kinetics which sirtuins are suited for substantial activation through base exchange relief.) For the mechanism-based inhibitor projects, we need to specify what advances our methodology brings to the table beyond the literature cited above. Are we proposing a computational approach for discovery of other leads like Ex-527, that could be used with any sirtuin? That could be novel, if Ex-527 and other leads like it were not discovered by computational lead discovery methods. Or are we proposing computational methods to optimize such leads? See above for comments on lead optimization.
RC (10-23): Both projects 1 and 2 should be split into two phases. Phase 1 - lead discovery. Phase 2 - lead optimization. We are dealing with Phase 1 now.
Project 1 (C pocket activator design):
Phase 1 - Develop LIA approaches for rapid prediction of Kd's of small molecules binding in C pocket. Computational advantages - backbone structural sampling is less important due to small molecules fitting into C pocket; molecules may fit into congeneric series. Inhibition mode validation via kinetic experiments. Show LIA model can provide reasonable Kd estimates for new molecules, noting that the experimental estimate for Kd may be somewhat inaccurate due to base exchange kinetics not being modeled in the inhibition kinetics equations (direct measurement of binding affinity may help). Experimental determination of extent of activation by the molecules; classification as inhibitors or activators. Target journal: PLOS CB?
Structural validation of docking protocol with known crystal structures: <list>
What is target range of Kd for this phase? (<to be filled in this week by PL/XG based on known Kd's of C pocket binders>)
PL(10/24): Project 1 is not about C pocket activator design. It is about examining the fitness of MM-GBSA, LIA, LIE in the lead discovery. The flow-chart in the presentation may be a little misleading, as I intent to apply MD in the complex validation stage (lead discovery), and not just limited to lead optimization. Assuming any of these method works reasonably well with a rigid receptor model (apo-enzyme or enzyme structural from complex) and selected ligands, we want to figure out why it works, and more importantly, for those doesn't fit to this model, if they can form some other series that work reasonably well together. If single receptor is not working for certain ligands, we can test if a single complex model as receptor may work for certain series (assuming we know the inhibition mechanism involving certain substrates). If the simple single-structure method is not working, can we implement some refinement steps (such as short MD (<1ns) + minimization) to get it work? If simple refinement is not working, can we implement long MD (>10 ns) simulation for refinement followed by MM-GBSA/LIE might work. We need a large collection of inhibitors for training and validation tests, and to come up with protocols depending on the lead discovery strategy and available structural and inhibition mechanism information. Currently we have some molecules tested in the lab, and there also exist some experimental data from literature, and I have come up a few methods to generate new leads, and some can be tested experimentally in training or validation.
RC: In that case, your project definitions may need to be modified. We will modify them here as required (see below), and will decide the precise interplay between experiment and computation in each project. Your input will be taken into consideration.
The salient feature of Project 1 as I defined it above is the focus on molecules that bind in a particular pocket so that they form a congeneric series, and the accurate prediction of binding affinities for these molecules (in the latter respect our definitions of the project are similar - accurate binding affinity prediction should certainly be a goal of project 1). Activators fall into this category because they bind in a particular pocket (C), but as noted, we do not restrict ourselves to activators in Phase 1. The reason to indicate activator discovery as a goal is that we know that binding affinity of C pocket binders will be lower than those of larger molecules like Ex-527, so maximization of binding affinity is clearly not the goal. By restricting ourselves to molecules that may fit into a congeneric series, we can interrogate the fitness of binding affinity models like LIA without sources of uncertainty associated with the receptor preparation procedure. Small molecules that fit in the C pocket will not require as extensive conformational sampling as larger molecules like Ex-527. C pocket binding by NAM and iso-NAM was also a focus of the 1st paper. Perhaps you feel that there are not enough molecules from the database that will fit into a single congeneric series to justify such an approach (e.g., Ex-527 was part of our initial experimental dataset, but we subsequently found that its mechanism of inhibition and effects on backbone conformation are quite different from the smaller molecules). We can discuss this.
A problem with the current subdivision of projects is that they have not subdivided the leads into classes that require similar approaches to conformational sampling. This may be a more systematic approach to project subdivision for the purpose of papers. Given that there are so many different types of mechanisms that fall into a given project as you defined it, a single project may not suitable for a single paper.
In the project subdivision you proposed, all modes of inhibition and all possible binding pockets were placed within the same project. I have indicated the possible problems with binding affinity prediction based on ensemble docking, which may be required for larger ligands. We need time to assess these issues and determine how MD can be used to resolve them. This could be done as part of a 2nd project.
It seems that you may be looking at projects from the perspective of computational workflow rather than papers - perhaps because you would like to try a variety of different approaches upfront to see which works and then settle which results fit into a paper. However, this approach may not be sufficiently well-organized to allow for proper project management and interaction between experiment and computation (as evidenced by the fact that we have not yet been able to prepare a detailed schedule of work for either one). If you would like to discuss these different perspectives toward project definition, we can arrange a time to meet.The detailed schedule of work for the first paper including both experimental and computational contributions is still due tomorrow (please aim to post earlier in the day so there time for discussion if needed).
Project 2 - sub-project 1 is about activator design as we are targeting the C pocket in the complex with intermediate in order to find a lead that can effectively blocks the base exchange reaction. Small molecules usually has a small binding affinity for sirtuins (in part due to the inability to overcome the entropy loss of flexible sirtuins), and will bind stronger with one or two substrates are bound as the protein already reduces its flexibility significantly. In this case, fast screening may be achieved using docking and MM-GBSA on single structure (probably with protein relaxation in the optimization step.) Activation effect can be tested experimentally. Also some of the small molecules may turn out work together with co-substrates to become an effective inhibitor.
Phase 2 - Base exchange kinetic experiments. Possible computational estimation of on/off rates of activator leads via MD. Possible use of QM/MM models for simulation of effects of activators on catalytic turnover (some of the info below on kinetic model can be copied to wiki page for this project). Show why certain leads are activators whereas others are inhibitors and establish the foundation for lead optimization of their potency. Possible issue: we may find after base exchange kinetic experiments that the particular sirtuin we are working with is not the best candidate for activation if base exchange inhibition is limited. Then consider using same leads with other sirtuins, and doing base exchange experiments on those sirtuins as well.
Target journal:
Detailed Phase 1 schedule needed this week from PL/XG.
Project 2 (inhibitor design; still requires subdivision into different modes of inhibition as noted above):
Phase 1 - Lead discovery of molecules that bind to pockets including but not limited to C, using ensemble docking, possibly induced fit, and some MD (see above for more details). Experimentally, identification of inhibition mode. More backbone structural sampling required. Possible issues w binding affinity calculations using ensemble docking. More potent inhibition possible. What is target range of Kd for this phase? (<to be filled in by PL/XG based on known Kd's of most effective sirtuin inhibitors that were discovered via screening without lead optimization.>)
Be aware that if our goal is lead discovery, the primary contribution may be a methodology for discovery of many such leads, since one might argue that there are already several promising leads like Ex-527 (which might have an easier time getting through clinical trials) and optimization is of these leads is more important than discovery of just one more lead that is no better than Ex-527. (Note there are not as many good activator leads.)
Target journal: PLOS CB?
Structural validation of docking protocol with known crystal structures: <list>
Novelty: <to be filled in this week by PL/XG>
Phase 2 - Lead optimization (binding affinity maximization) via FEP and mutations of drug molecule leads. Possible collaboration/interaction with Schrodinger/Nimbus/DE Shaw colleagues regarding lead optimization methods. Before embarking on this phase, we must establish that a CRO can successfully solve the crystal structure of one of our lead molecule-sirtuin complexes, so we can continue solving crystal structures during the lead optimization process.
Target journal:
PL(10/24): Project 2 is mostly about lead discovery of inhibitors. There are different inhibition mechanisms, i.e. Analogues of NAD+ and Kinase inhibitors (Ro31-8220, GW5074, etc) competitively vs NAD+; Nicotimamide analogues occupied C pockets and also extend to the acetylated peptide binding pocket, and inhibits competitively vs acetylated peptide. Cambinol works the same way, and probably hydroxynaphthaldehyde and thiobarbiturate inhibitors; Ex-527 have decent binding affinity when NAD+ exists, but it works mostly through binding together with co-product (2'-OAADPR), not intermediate that we originally thought. There are also inhibitors works together with the ATP end of NAD+ in the A pocket and nicotinamide moiety in other position (SRT1720 in SIRT3, and Ex-527). Suramins works noncompetitively with both NAD+ and peptide substrate. (The survey is still going on.)
Currently, most computational drug discovery focus on receptor-based virtual screening, that finds best binding to apo-enzyme, that corresponds to competitive inhibition with one or both substrates. There are also groups discover new leads through identifying unique scaffolds. However, if we focus on the inhibition mechanism, we can discover new leads effectively. Having a co-substrate such as NAD+ or peptide substrate or products in place may be easier than competing with both substrates. I have recently identify a small molecules that binds reasonably well to the C pocket (based on Glide XP and MM-GBSA calculation), and we may use it as a starting block to grow/find new leads (for comparison ELT inhibitors grow from thieno[3,2-d]pyrimidine-6-carboxamide).
RC: There are too many modes of inhibition considered under one project. As I mentioned, answering in paragraph form is not efficient (e.g., please subdivide the inhibitors into classes in list form). I am ok with focusing on lead discovery in this project, rather than binding affinity prediction (in that case, perhaps J Med Chem would be more appropriate), but there is clearly overlap between the two projects as you have defined them, and this overlap would not exist if the projects were subdivided by inhibition mechanism/binding mode. We all agree that mechanism-based discovery is important (given that our first paper was on the inhibition mechanism), but the key is to enumerate the possible mechanisms and establish a detailed plan for discovery associated with each. The methods used to dock and accurately predict binding affinities may differ for each class. Each such class may have a different target Kd.
Please make wiki pages for these projects, renaming existing wiki pages if needed to avoid confusion (e.g. page titles like "Docking Simulations" may be ambiguous going forward).
RC: Equipment needs - Experiment: background work needs to be done to get radioactive assay approval, some additional equipment quotes including those for binding affinity assays. Computation: benchmarking use of multiple compute nodes for MD simulations.
RC: Regarding explicit waters, note that there are several methods (including watermap) that are used in the context of docking (e.g., Nimbus uses these routinely for lead discovery and optimization). We mentioned issues with explicit waters and need for predicting their positions in one version of our 1st sirtuin paper. A methodology/tool should be chosen for placement of explicit waters during lead discovery.
Group meeting on Sirtuin Project 10-19-2013
XG(10/19): Presentation on experimental part. group meeting_101813.pptx
PL(10/19):
Here is my presentation for the group meeting. Groupmeeting_SIRT3_10-19-2013.pptx
Here is some of reference collection in BibTeX format. PL_reference_Collection_10-19-2013.bib
Xiangying and I will discuss in more details the planed work flow (with some estimates on the time) for each specific cases of inhibitors/activators discovery, both computationally and experimentally.
RC: When you get a chance, please also provide some more info on ensemble docking and how one can hope to obtain binding free energies with this technique, given that the starting structures used for docking different drugs will be different, and MD sampling will not be sufficient to erase memory of the different starting structures used to dock these drugs.
PL: Here are two review articles by J Andrew McCammon and his colleagues on the computational techniques for drug-discovery. 1741-7007-9-71.pdf 1-s2.0-S1471489210001372-main.pdf and this review article discuss more about how to combine docking and MD in drug design. Alonso, H., Bliznyuk, A. a & Gready, J. E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 26, 531–68 (2006)
It is important to take into account of protein flexibility, and ensemble docking is one way of addressing this issue. Some of the ensemble docking use MD generated conformations as receptor pool. Any single receptor structure is biased toward certain set of ligands. Currently ensemble docking is mainly used in capturing the correct pose (not final bound structure). Once the correct pose is identified, we can use a sufficiently long MD and maybe some local enhanced sampling to refine the structure and get a good sampling. Therefore, we can at least provide a way to correct some of the bias. If the docked pose is closer to its native bound structure, we have a better chance of using MD to obtain more accurate binding free energy.
RC: Thanks. Before embarking on a project that aims to compute binding free energies starting from an crystallographic ensemble of structures, we will need to have a more detailed analysis, possibly based on this literature, for whether we can in fact use MD sampling with ensemble docking in order to get reasonably accurate binding free energies. Otherwise, we may run into problems writing a paper after many months of work.
I can see that if the ensemble is generated by MD, this will not be a problem. At least for congeneric series, we can dock to all the pdb structures above, but then ultimately use just one starting structure for the binding affinity comparisons. I can also see how ensemble docking can be a very useful tool for
Overview of the Sirtuin Project and General Notes
XG(8-16): The reported sirtuin deacetylation catalytic mechanism, including the experimental kinetic parameters, methods and computitional calculations using QM/MM, has been collected. Not finished.
Catalytic mechanism of Sirtuin Deacetylase Reaction_Computational calculation_ experimental methods.pptx
The QM/MM calculation related references are uploaded. Not finished.
Investigation of the catalytic mechanism of sir2 enzyme with AMMM approach_2010_Liang.pdf
Improved pseudobonds for combined ab initio quantum mechanical_2005_Zhang.pdf
Supp_Investigation of the catalytic mechanism of sir2 enzyme with AMMM approach_2010_Liang.pdf
Supp_Highly dissociative and concerted mechanism for the nicotinamide_2008_Hu.pdf
Highly dissociative and concerted mechanism for the nicotinamide_2008_Hu.pdf
A pseudobond approach to combining quantum mechanical and molecular mechanical methods_1999_Zhang.pdf
RC: Thanks - are there any reports of on/off rates of NAD+ or NAM, or methods for determining them?
XG(8-19): Some of the rate constants for different steps of deacetylation reaction were measured experimental in papers. For example (see follwoing picture), k4 and k5 discrible the detailed measurement for NAM exchange reaction. Noticed that, most of the work is done on Sir2Tm.
RC (11-4): Revisiting this model since it is relevant for our current project: please confirm that the base exchange binding of NAM is studied only experimentally, through MM kinetics, and not computationally. NAM binding is modeled using Km in the MM formulation, and the on-off rates (if desired, as they may be for a complete kinetic model suitable for arbitrary substrate and inhibitor concentrations, not just high concentrations) can be determined through some additional experiments that you outlined below. Please also confirm that kcat for base exchange was computed by QM/MM and also measured experimentally through MM kinetics. Were the computationally predicted and experimentally determined values compared? What rate constants if any were determined solely through QM/MM? I assume these were all catalytic rate constants?
Regarding inhibition kinetics for the base exchange reaction, please confirm that NAM is treated as the substrate here and that isoNAM would act as a competitive inhibitor of the base exchange reaction, thereby activating the deacetylation reaction. So modeling of the Ki of isoNAM (or other C pocket binders) according to the conventional inhibition models requires one to specify whether one is considering inhibition with respect to the substrate NAD+ or with respect to the substrate NAM. But typically, when one is determining Ki's for C pocket binders, one works at sufficiently low NAM concentrations that the effect of base exchange on deacetylation rate is negligible. Please confirm. However, when we model activation, this effect cannot be neglected; high concentrations of both NAM and NAD+ are used and the full kinetic model (rather than a MM model of either the deacetylation or exchange reaction) is used to predict the effect of the activator on the overall deacetylation rate. Note however that without a way of determining the on/off rates of a potential activator, we may have no choice except to resort to a MM model that assumes a steady state value of the concentration of the bound activator computed via the associated Ki.
Ki in the table below refers to that of NAM.
sauve.pdf
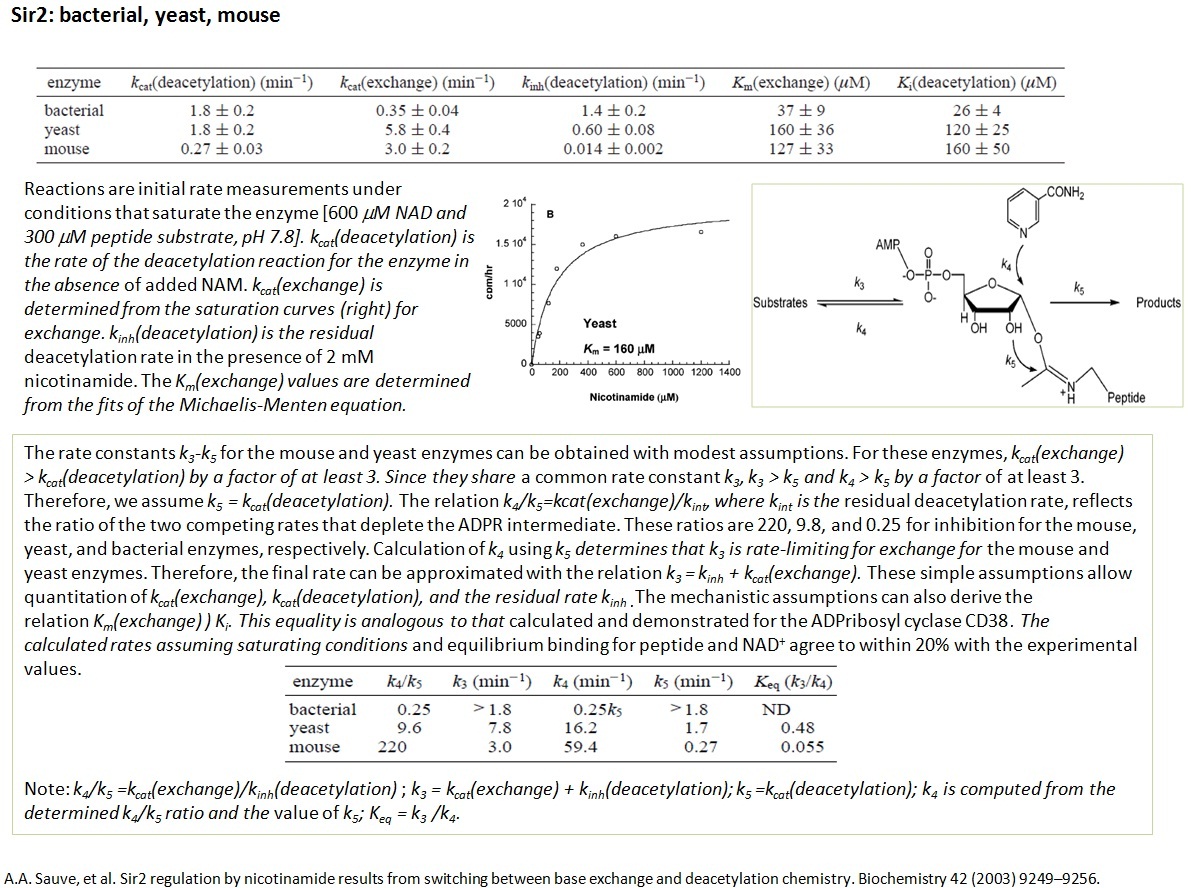
The literature search is still on. Will have a whole picture when it is done.
RC (8-22): Can you clarify whether k4 is a binding rate constant or does it include a contribution from kcat (i.e., is it of the form of a 2nd order rate constant kcat/Km). Also, do you know what methods people use to determine such on-off rates?
XG(8-23): In base exchange reaction, NAM works as a exchange substrate. k4 is a binding rate constant.
To measure k4, several experiments need to be carried out.
Exp I_kcat(deacetylation): Measure the rate of deacetylation reaction without addition of NAM.
Exp II_kinh(deacetylation): Measure the residual deacetylation rate in the presence of 2 mM of NAM.
Exp III_kcat(exchange): Measure the rate of exchange reaction in the presence of different concentration of [carbonyl-14C]nicotinamide.
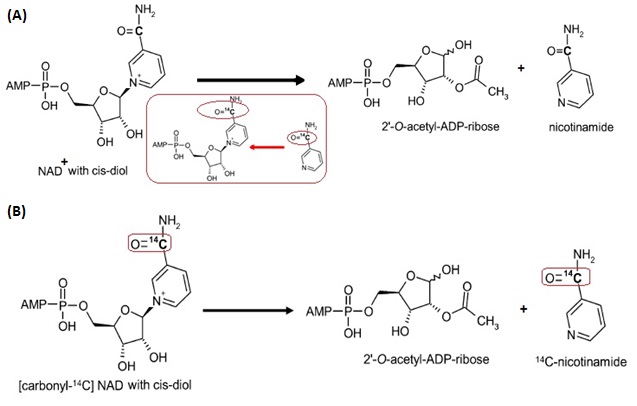
At the mean time, ADPR (intermediate) and 2’-O-acetyl-ADPR products need to be measured. The production of these compounds is stoichiometrically linked with lysine deacetylation and can be used to quantify deacetylation.
RC(8-22): It seems the existing literature data is providing a solid foundation for our proposed simulations of the reaction/activation kinetics. We will see what data is missing and whether computational methods can help.
XG(8-23): Yes. All the reported data are for Sir2Tm. We can apply to other members in Sirtuin family.
RC (8-8):
We can start setting up a kinetic model for sirtuin catalysis based on all the rate constants that have been determined in
previous literature using QM/MM and experiments. There will be some missing rate constants, like the on/off rates of the drugs. We can then
explore through simulations (somewhat like simulations we are doing for the PCR reaction network) what types of on/off rates for the drugs would
lead to efficient activation. This would help establish viability of our planned work on activator discovery.
XG(8-8): Good idea! You open another window for sirtuin research and also make a crosstalk among projects. Will do.
RC (8-7):
.
We have been reviewing recent literature on sirtuins. I have a few comments on these vis-a-vis the plans for our work:
1) The Lin papers that got into Science and Nature follow a certain pattern: they explore features of the wild-type enzyme (like substrate specificity) in the absence
of any modulators. Substrate specificity is a natural topic to study for a chemical biologist like Lin - the work involves synthesizing new substrates and high-throughput assaying, which are things within their domain of expertise. I agree computation can help here: one adds functional groups to the peptide substrate and checks on the computer what types of functional groups fit in the binding pocket with reasonable binding affinity. Since one is often looking at side chain modifications to a peptide, it may not be necessary to dock the modified substrate from scratch.
Science and Nature like these papers if they broaden our understanding of important wild-type enzymes. Lin has chosen high-value enzymes for his application of standard chemical biology tools. It is harder to get a paper into Science or Nature based on design of modulators. However, we are more interested in the latter goal (though we may study modulation of deacetylation of new substrates of SIRT5 and 6).
2) I'm not surprised that SIRT3 inhibitor discovery papers are not using some of the advanced binding affinity estimation tools. Our second paper should be novel in this regard. If you haven't already done so, perhaps you could collect all the Sir2 computational binding affinity prediction papers Eric looked at so the new hire can study them.
Inhibitor discovery: we will be using LIA to train computational MM-GBSA models for prediction of binding affinities of new inhibitors (lead discovery). The leads can subsequently be optimized using more computationally intensive MD calculations. After predicting a lead and then verifying it experimentally, we will then do MD calculations to see if we can more accurately reproduce its experimentally observed binding affinity. Then by looking at MD trajectories, we will try to determine what modifications will facilitate even tighter binding.
3) QM/MM: I looked at papers on QM/MM modeling of the Sir2 catalytic cycle. The value of these studies for us is in the role they can play in -activator discovery-.
QM/MM techniques can provide estimates of rate constants in multistep catalytic reactions that are difficult to obtain experimentally. Moreover, even if they can be obtained experimentally, QM/MM can provide insights into what residues are important in lowering energetic barriers to reaction.
--For us, the most important QM/MM study is to investigate (possibly in more than one sirtuin) the energetics of base exchange--. This should allow us to explain the branching ratio between the formation of product and reversal of reaction after step 1 (NAD+ cleavage) in terms of the respective rate constants. The branching ratio will determine the potential for activation of a given sirtuin. -This QM/MM study could be done routinely by the new hire at an appropriate time, starting from prior published work.- Simultaneously, we may apply your new assay to experimentally determine the branching ratios.
Activation will first be studied in the case of iso-NAM. If we can obtain estimates of the important rate constants in the catalytic cycle, we can predict through simulation of the reaction network the effect of an activator on the catalytic efficiency of a sirtuin. Important rate constants here include the on/off rates of both NAM and iso-NAM. Sometimes one cannot easily measure these rates in experiments. We will need to determine which of the rates can be experimentally measured. MD calculations can sometimes provide decent estimates of these rate constants. These on/off rate constants can be combined with the branching ratio rate constants for the catalytic steps studied using QM/MM to simulate the effect of the activator on efficiency.
*Related, please let me know what experimental techniques can be used to experimentally determine the on/off rates of inhibitors and substrates.
MD studies could reveal why iso-NAM is a good activator, based on what structural features lead to it having appropriate on/off rates for good activation. Then we can connect up with paper 2) by taking the lead compounds discovered there as a starting library for identification of activators. These leads can be refined by comparing MD simulations to MD simulations of iso-NAM. If we can identify new activators by this approach, the paper could go in a high profile journal because of its explanation of how one can rationally design activators.
Activation by protein engineering: Here the computational scientist will be developing codes for high-throughput sampling of mutant protein sequences that can bind tightly to NAD+ but bind only weakly to NAM, allowing NAM to dissociate quickly. This is connected to Chaoran's beta-lactamase project. MD can be used on the top-ranking leads to refine the predictions of their binding affinities for comparison to experiment and lead optimization. QM/MM and MD could be used to determine what residues are ok to mutate; they could also be applied to the engineered enzymes, if they are able to increase activity.
The above plans outline several possible roles of the computational scientist on the sirtuin project. Note that the activation studies are focused on the concept of activation by derepression. It is important to first identify the potential for activation by looking at branching ratios before proceeding too far with this.